ANN - Classification: Diabetes Prediction
Learning Goals
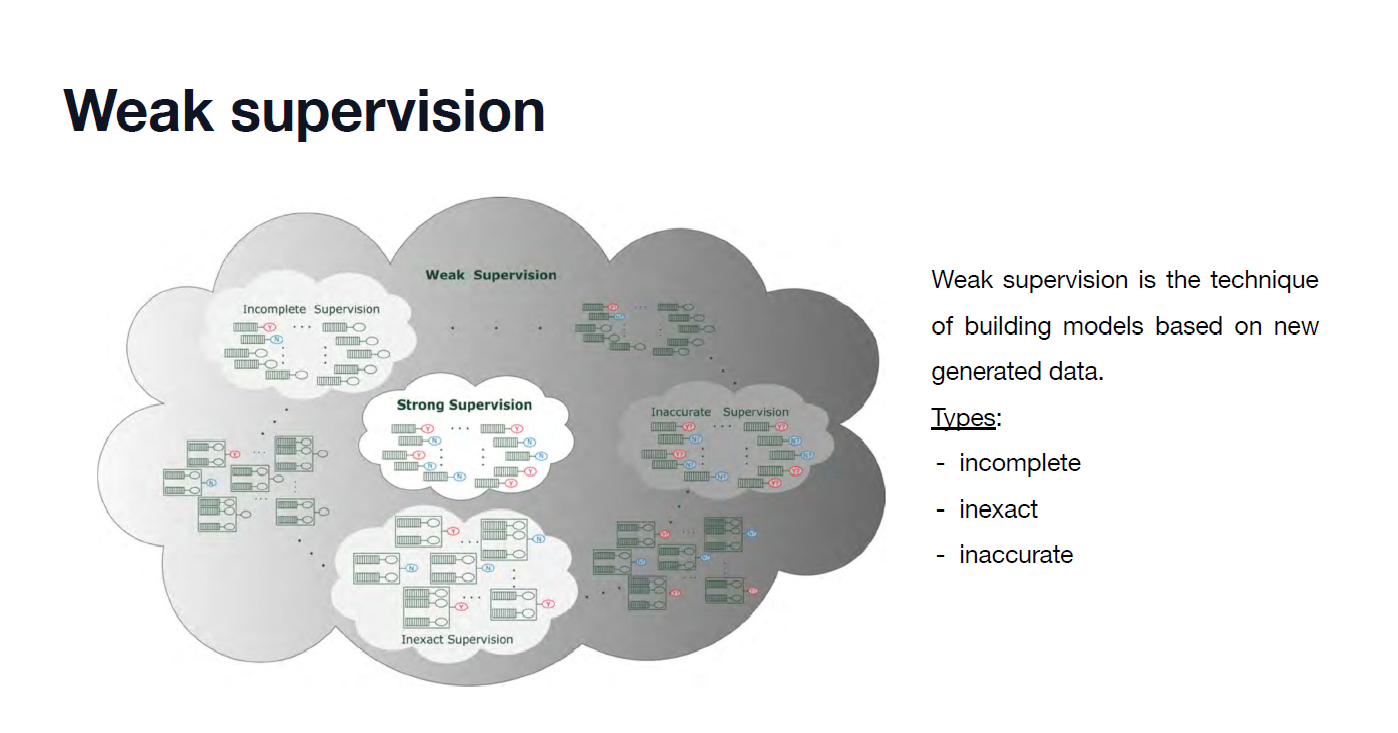
ANN을 이용한 당뇨병 예측 모델 구축 Diabetes prediction model using ANN
Loading the dataset
[Notice] Download Dataset (Kaggle)
import tensorflow as tf
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
diabetes = pd.read_csv("diabetes.csv")
diabetes.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 768 entries, 0 to 767
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Pregnancies 768 non-null int64
1 Glucose 768 non-null int64
2 BloodPressure 768 non-null int64
3 SkinThickness 768 non-null int64
4 Insulin 768 non-null int64
5 BMI 768 non-null float64
6 DiabetesPedigreeFunction 768 non-null float64
7 Age 768 non-null int64
8 Outcome 768 non-null int64
dtypes: float64(2), int64(7)
memory usage: 54.1 KB
diabetes.columns
Index(['Pregnancies', 'Glucose', 'BloodPressure', 'SkinThickness', 'Insulin',
'BMI', 'DiabetesPedigreeFunction', 'Age', 'Outcome'],
dtype='object')
sns.countplot(x = "Outcome", data = diabetes)

당뇨가 있다면 1, 없으면 0이다. 1 if diabetes is present, 0 otherwise.
sns.heatmap(diabetes.corr(), annot = True)

한 눈에 보기 좋게 변수들 간 상관관계를 히트맵으로 표시했다. At a glance, correlations between variables are displayed in a heat map.
X = diabetes.iloc[:, 0:-1].values
y = diabetes.iloc[:, -1].values
X.shape, y.shape
(768, 8) (768,)
독립변수는 8개, 종속 변수는 output 한 개만 존재한다. # independent variables: 8, # dependent variables: 1
Feature Scaling
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X = sc.fit_transform(X)
종속 변수는 당뇨병 유무로 0 or 1이기 때문에, scaling이 필요없다. Since the dependent variable determines whether a patient has diabetes or not (0 or 1), it doesn’t require scaling.
Splitting the dataset into the Training set and Test set
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2)
Building the model
classifier = tf.keras.models.Sequential()
classifier.add(tf.keras.layers.Dense(units=400, activation='relu', input_shape=(8, )))
classifier.add(tf.keras.layers.Dropout(0.2))
classifier.add(tf.keras.layers.Dense(units=400, activation='relu'))
classifier.add(tf.keras.layers.Dropout(0.2))
classifier.add(tf.keras.layers.Dense(units=1, activation='sigmoid'))
classifier.summary()
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense (Dense) (None, 400) 3600
dropout (Dropout) (None, 400) 0
dense_1 (Dense) (None, 400) 160400
dropout_1 (Dropout) (None, 400) 0
dense_2 (Dense) (None, 1) 401
=================================================================
Total params: 164,401
Trainable params: 164,401
Non-trainable params: 0
_________________________________________________________________
뉴런 개수는 임의로 설정한 값이므로 수동으로 바꿔가며 최적값을 찾을 필요가 있다. Since the number of neurons is an arbitrarily set value, it is necessary to find the optimal value by manually changing it.
classifier.compile(optimizer='Adam', loss='binary_crossentropy', metrics = ['accuracy'])
epochs_hist = classifier.fit(X_train, y_train, epochs = 200)
Epoch 1/200
20/20 [==============================] - 0s 4ms/step - loss: 0.1085 - accuracy: 0.9658
Epoch 2/200
20/20 [==============================] - 0s 4ms/step - loss: 0.1234 - accuracy: 0.9446
Epoch 3/200
20/20 [==============================] - 0s 4ms/step - loss: 0.1201 - accuracy: 0.9577
...
Epoch 199/200
20/20 [==============================] - 0s 5ms/step - loss: 0.0146 - accuracy: 0.9951
Epoch 200/200
20/20 [==============================] - 0s 5ms/step - loss: 0.0135 - accuracy: 0.9967
y_pred = classifier.predict(X_test)
y_pred
array([[1.91377112e-05],
[3.52401704e-01],
[7.71102607e-01],
[8.95466566e-01],
[9.94010568e-01],
[2.07010522e-01],
...
[1.37080904e-02],
[1.79921073e-04],
[3.26797456e-01],
[8.01205460e-05],
[4.79539931e-02]], dtype=float32)
상기 수치들은 각각의 시행에 대하여 당뇨병이 존재할 확률을 의미한다. The above figures represent the probability of the presence of diabetes for each trial.
만약, 그 확률이 절반 이상일 경우 당뇨가 있다고 가정해보자. If the probability is more than half, let’s assume that you have diabetes.
y_pred = (y_pred > 0.5)
y_pred
array([[False],
[False],
[ True],
[ True],
[ True],
[False],
[ True],
...
[False],
[False],
[False],
[False],
[False]])
Evaluating the model
epochs_hist.history.keys()
dict_keys(['loss', 'accuracy'])
plt.plot(epochs_hist.history['loss'])
plt.title('Model Loss Progress During Training')
plt.xlabel('Epoch')
plt.ylabel('Training and Validation Loss')
plt.legend(['Training Loss'])

손실 분포의 분산을 낮추려면, batch 혹은 뉴런 개수를 늘리면 된다. To lower the variance of the loss distribution, you can increase the number of batches or neurons.
하지만 이렇게 할 경우, 학습 속도가 느려진다. However, doing so slows down learning.
대신, 모델 정확도가 올라가서 학습 데이터의 오분류 개수가 줄어들 수 있다. Instead, the number of misclassifications in the training data may be reduced by increasing the model accuracy.
하지만, 과적합 가능성 또한 증가한다. However, the possibility of overfitting also increases.
모델 학습에는 이러한 trade-off 관계가 존재한다. This trade-off relationship exists in model training.
보다 자세한 내용은 SGD vs. Mini-Batch vs. BGD를 참조하자. For more information, see SGD vs. Mini-Batch vs. BGD.
Confusion Matrix
from sklearn.metrics import confusion_matrix
y_train_pred = classifier.predict(X_train)
y_train_pred = (y_train_pred > 0.5)
cm = confusion_matrix(y_train, y_train_pred)
sns.heatmap(cm, annot=True)

Classification Report
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
precision recall f1-score support
0 0.78 0.74 0.76 102
1 0.53 0.60 0.56 52
accuracy 0.69 154
macro avg 0.66 0.67 0.66 154
weighted avg 0.70 0.69 0.69 154




댓글남기기